RecA Protein

|
Table of Contents:
- Introduction
- Structure
- Function
- Disease
|
Introduction to the RecA protein
RecA is a protein that catalyzes the genetic processes of general recombination and DNA repair. Its eukaryotic equivalent is the Rad51 protein.
- General recombination is a process of genetic exchange that occurs across homologous sequences of DNA. The homologs of two chromosomes are aligned according to the homologous sequences shared between them. The single strands of these identical homologs are then broken to create recombinogenic ends. An RecBCD complex binds to the ends and exonucleases (cuts) the double stranded DNA until it comes across a chi site. This chi site inhibits the continued slicing of the 3' end. (www.ncbi.nlm.nih.gov) This now allows for strand invasion where the exposed 3' end of one homolog inserts itself between the double strands of the other identical homolog thereby displacing one of the original strands. The resulting Holliday junctions are later resolved by RuvC endonuclease which cuts the junctions and binds the DNA resulting in two separate chromosomes that have been recombined. (www.ncbi.nlm.nih.gov)
* Helpful animation of general recombination
- DNA repair is a process used to repair and correct damages in DNA sequences. DNA repair can be facilitated by several different mechanisms including direct reversal, Nucleotide Excision Repair, DNA Mismatch Repair, Double Stranded Break Repair, and other post replication repairs (www.mun.ca/biochem). RecA has a critical role in Double Stranded Break Repair as well as in post replication repairs, specifically the prokaryotic SOS Response.
RecA Structure
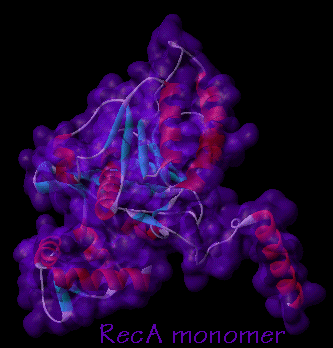
- A RecA monomer consists of three domains: (www.callutheran.edu)
* a large, central domain
* an amino domain and
* a carboxy domain
* The central domain functions to faciliate DNA and ATP binding. (www.callutheran.edu)
* The amino domain is important in the forming the RecA polymer. (www.callutheran.edu)
* The carboxy domain assists in interfilament associations. (www.callutheran.edu)
- These RecA monomers can come together to form a RecA filament.
* The RecA filament utilizes an amino to central domain polarity that binds RecA monomers together forming the filament. (www.callutheran.edu)
* The filaments create a helix around the single-stranded DNA. (www.callutheran.edu)
> Consists of 6 RecA monomers per revolution
> Contains ATP within the helical center
Function of RecA
- The RecA protein's main function is within the process of general recombination. RecA has two binding sites within its structure - one for the single stranded DNA and one for the duplex of the double stranded DNA. (Heerssen) RecA causes the single strand to straighten and extend into the double strand helix creating a triple strand formation. The RecA binding sites hold the single strand in place while also allowing the double stranded duplex to unwind. As this occurs, the single strand searches for its homologous sequence in the unwinding double stranded DNA. (Heerssen)
- By binding to the triple strand formation, RecA has initiated strand exchange, created a heteroduplex, and facilitated branch migration.
* Another helpful animation for understanding RecA's role in general recombination
- RecA is also crucial in postreplicative repair like the prokaryotic SOS Response. The SOS Response activates mulitple genes that allow DNA replication to overlook errors. This response is especially helpful in the survival of Ultra-Violet damaged cells. When a cell is damaged, by UV light for example, the cell initiates its SOS Response. (Ptashne) RecA is called in to act as a protease that cleaves the lambda repressor and LexA. LexA is a repressor that normally inhibits proteins such as uvrA and uvrB that aid in the repair of UV damaged DNA. (Ptashne) RecA binds with LexA on the DNA and forces it to self-cleave. RecA also cleaves lambda repressor monomers. This decreases the amount of lambda repressor dimers being formed, decreases the amount of bound Or2 and Or1, and decreases the transcription of the repressor. This initiates Cro expression which allows the cell to undergo lytic growth and lysis. (Ptashne)

RecA role in disease
- RecA has been studied for its role in cloning and genetic engineering. (wikipedia.com)
* It was found that strands of E.coli that were lacking in RecA monomers allowed plasmids inserted into DNA to be left unaltered and able for retrieval.
> This property could be an effective tool for cloning procedures. (wikipedia.com)
* RecA protein has also been utilized to change DNA constructs and aid in the genomic DNA mapping.
- Antibiotic resistance is continuing to increasingly spread worldwide posing a serious medical threat.
* Tim Wigle and Scott Singleton of University of North Carolina School of Pharmacy have aimed their research towards finding ways to stop the growing antibiotic resistance crisis. Their research focuses on the development and transmision of microbial drug resistance and its control. Through their research they have identified RecA as a vital component of both the spread and development of resistance. As bacteria become stressed by antibiotics they trigger the RecA protein which in turn signals the SOS Response. The SOS Response initiates the transcription of the bacteria and thereby allows it to undergo lytic growth and survive with thousands of replications. If an SOS Response is not initiated, the RecA protein facilitates mutation of the bacteria that allows it to survive the antibiotic stress. The recombinogenic nature of RecA also initiates the bacterial DNA to undergo strand invasion which facilitates the sharing of this now resistant bacterial DNA. (www.pharmacy.unc.edu)
Wigle and Singleton are currently searching for RecA inhibitors that will hopefully act to slow down the bacterial transcription rate or make the bacterial DNA more susceptible to current antibiotic drugs. So far no effective compounds have been found. (www.pharmacy.unc.edu)
* Matthew Waldor, John Beaber and Bianca Hochhut, Howard Hughes Medical Institute researchers, have also been studying the effects of RecA on antibiotic resistance. This research group has focused on a particular set of antibiotic-resistant genes, SXT which currently resistant to four different antibiotics. SXT contains setR which represses setC and setD, activators of the excision and transfer of SWT. So under normal conditions setR maintains SXT in an inactive state. However, the presence of RecA inactivates setR by proteolysis. RecA initiates the SOS Response which activates setC and setD allowing transfer of the SXT gene. A major finding within their research was that this SOS response and resulting resistance is initiated not only by UV light damage but also by the presence of certain antibiotics such as ciprofloxacin. (www.hhmi.org)
References
Cox, M.M. The Bacterial RecA Protein as a Motor Protein. Annual Review of Microbiology, 2003. Online <http://arjournals.annualreviews.org/doi/abs/10.1146/annurev.micro.57.030502.090953?cookieSet=1&journalCode=micro>.
Ferrin, L.J. Flexible Genetic Engineering Using RecA Protein. DNA Repair Protocols: Prokaryotic Systems, 2000. Online <http://www.springerprotocols.com/Abstract/doi/10.1385/1-59259-068-3:135>.
Fighting the Resistance: Wigle works to inhibit growth of antibiotic-resistant bacteria. UNC Chapel Hill, 2009. Online <http://www.pharmacy.unc.edu/experience/the-life-of-a-unc-sop-phd/research-highlights/tim-wigle>.
Heerssen, H.M., D. Marcey, and A. Downs. The RecA Protein: Structure and Biological Function. 2001. Online
<http://www.callutheran.edu/Academic_Programs/Departments/BioDev/omm/reca/recamast.htm>.
Ptashne, M. A Genetic Switch: Third Edition Phage Lambda Revisited. Cold Spring Harbor Laboratory Press, New York. 2003.
RecA Protein, E. coli. Epicentre Biotechnologies. 2009. Online <http://www.epibio.com/item.asp?ID=374>.
Researchers Identify New Mechanism Behind Spread of Antibiotic Resistance. HHMI News, 2003. Online <http://www.hhmi.org/news/waldor.html>.
Comments (0)
You don't have permission to comment on this page.